SQL Server (and also Azure SQL Managed Instance) has built-in PREDICT() function and Machine Learning Services component, which enables you to work with machine learning workloads (such as, running R or Python scripts) in SQL Server process without transferring data across network.
In short, you can run your ML workloads close to data.
You can take several approaches (patterns) for running in-database ML workloads.
Here I summarize these patterns and show you several things you should know about.
Our Sample (Dataset)
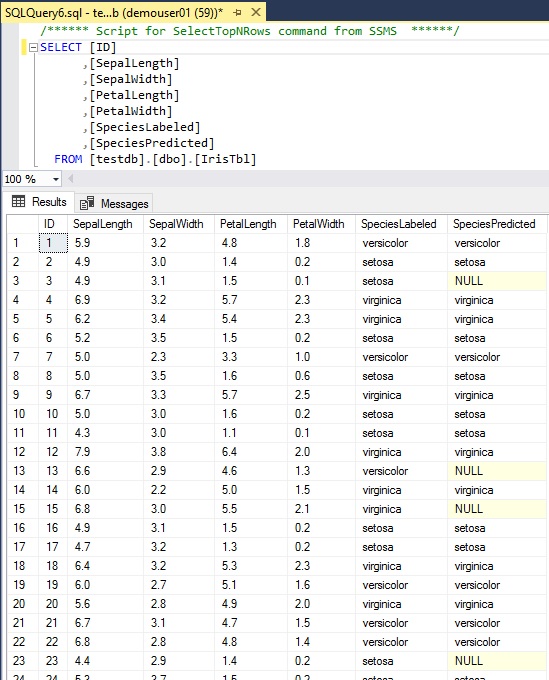
In this post, we use the following “IrisTbl” table, which is the famous iris dataset containing 3 classes of iris species with several features (sepal length, petal width, etc), in SQL Server database. (You can download from here.)
This dataset has 150 records. Among these rows, 130 records have been already predicted for the class of species and this predicted value is in “SpeciesPredicted” column. For other 20 rows, the class of species is not still predicted and “SpeciesPredicted” values are all null. We’ll predict the class for these 20 rows.
When the new record is added in this table, the “SpeciesPredicted” column will be scored (predicted) in batch.
This table can also be used in training.
All rows are having “SpeciesLabeled” column, which is used for label (i.e, actual value). We’ll train a model using the already predicted rows (130 rows), since we’ll predict other rows (20 rows) using the trained model.
We can also use this label (“SpeciesLabeled” column) for seeing whether the predicted value is correct or not.
In this post, we’ll use R script, but you can also run the same logic with Python script.
Pattern 1 : R / Python Scripting in Stored Procedures (sp_execute_external_script)
The first approach is to use sp_execute_external_script stored procedure, which enables you to call R or Python script running on SQL Server Machine Learning Services.
Using this approach (and next approach), you can run your ML workloads in database objects, such as, database triggers, database transactions, batching your stored procedures with SQL Server Agent, and integrating with SQL Server management utilities (resource governor, and so on).
Note (Oct 2018) : In addition to R and Python, now Java (JVM) is supported in SQL Server ML Services as language extension. For instance, you can serialize your Spark ML pipeline model as MLeap bundle and bring it into SQL Server.
Let’s see the following sample code.
Here we call R script in SQL stored procedure, in which we train the model with decision tree algorithm. This stored procedure returns the generated model binary (which type must be VARBINARY(MAX)) as one of output arguments.
CREATE PROCEDURE GenerateTreeModel ( @model_name VARCHAR(50) OUTPUT, @model_data VARBINARY(MAX) OUTPUT)ASBEGIN EXEC sp_execute_external_script@language = N'R',@script = N'model <- rxDTree( SpeciesLabeled ~ SepalLength + SepalWidth + PetalLength + PetalWidth, data = IrisData, cp = 0.01)modelname <- "Decision Tree"modeldata <- as.raw(serialize(model, connection=NULL))',@input_data_1 = N'SELECT * FROM IrisTbl WHERE SpeciesPredicted IS NOT NULL',@input_data_1_name = N'IrisData',@params = N' @r_rowsPerRead int, @modelname varchar(50) OUTPUT, @modeldata varbinary(max) OUTPUT',@r_rowsPerRead = 10000,@modelname = @model_name OUTPUT,@modeldata = @model_data OUTPUT;END;GO
As you can see above, you can pass data between SQL script and R script using input / output parameters. (In this example, we only use one single input parameter.) When you don’t define these parameters, the default value – which name is “InputDataSet” for the input parameter and “OutputDataSet” for the output parameter – is used.
Now let’s invoke this stored procedure (GenerateTreeModel) as follows.
The following code is saving a generated model (a trained model) into “ModelTbl” table. As you know, this procedure runs on SQL Server process and the generated artifacts should then be stored in database. (You cannot save this model as a regular file.)
-- model table definitionCREATE TABLE ModelTbl ( ModelName VARCHAR(50), ModelData VARBINARY(MAX) );GODECLARE @model_name VARCHAR(50), @model_data VARBINARY(MAX);-- Invoke GenerateTreeModelEXEC GenerateTreeModel @model_name OUTPUT, @model_data OUTPUT;-- Save model in tableIF EXISTS(SELECT * FROM ModelTbl where ModelName=@model_name) UPDATE ModelTbl SET ModelData=@model_data WHERE ModelName=@model_nameELSE INSERT INTO ModelTbl(ModelName, ModelData) VALUES(@model_name, @model_data);
One important note is : With SQL Server Enterprise Edition, data can be streamed and scaled with parallelization.
On contrary, this scaling is not available with other editions and eventually the workloads will fit into memory. If data is so large, the allocation failure will occur with these editions.
Note : The actual number of execution processes depends on several factors (server resources, and so on).
With this reason, it’s better to use RevoScaleR or MicrosoftML functions (revoscalepy or microsoftml in python), even though you can use standard R functions in sp_execute_external_script. Even when your server is having sufficient RAM, the default size of external resources on SQL Server is limited for not preventing database workloads. Eventually you might suffer for running large training tasks without RevoScaleR (or MicrosoftML) functions. (See here for increasing external resource size.)
By using RevoScaleR functions, the procedure is optimized by multiple process. Data is read by chunk, and the operation is also distributed in multiple processes. The results are eventually aggregated. (See my early post “Benefits of Microsoft R and Machine Learning Server (R Server)” for RevoScaleR package.)
In the later section in this post, I’ll show you another approach to use SQL Server Compute Context. If you use RevoScaleR functions, you can also use SQL Server Compute Context instead of using sp_execute_external_script.
Note : You can install additional packages using RGui tool in SQL Server. See the official document “Install additional R packages on SQL Server” for details.
[Added on Oct 2018] Now you can easily manage (install, list, delete) packages using new sqlmlutils library.Note : When you want to parallelize with standard R functions in
sp_execute_external_scriptstored procedure, you must explicitly set@parallelparameter (parallelize or not) insp_execute_external_scriptby yourself. (When you use RevoScaleR functions, this parameter is automatically set.)
You can also use sp_execute_external_script for prediction (scoring).
The following code retrieves a trained model (binary) from table, and predicts the label using this model.
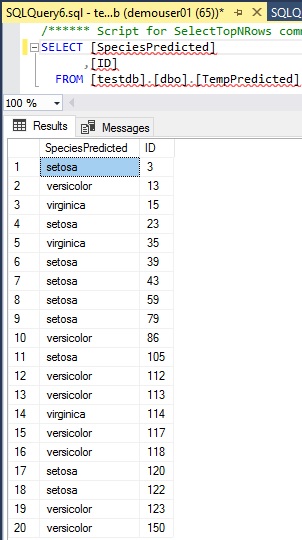
CREATE PROCEDURE GetPredictedResultsAsTable ( @modelname VARCHAR(50))ASBEGIN DECLARE @model_data VARBINARY(MAX) = (SELECT ModelData FROM ModelTbl WHERE ModelName = @modelname); DROP TABLE IF EXISTS TempPredicted; CREATE TABLE TempPredicted (SpeciesPredicted VARCHAR(50),ID INT ); INSERT INTO TempPredicted EXEC sp_execute_external_script@language = N'R',@script = N'model <- unserialize(modeldata)OutputDataSet <- rxPredict( model, data = IrisScoreData, type = "class", predVarNames = "SpeciesPredicted", extraVarsToWrite = c("ID"))',@input_data_1 = N'SELECT ID, SepalLength, SepalWidth, PetalLength, PetalWidth FROM IrisTbl WHERE SpeciesPredicted IS NULL',@input_data_1_name = N'IrisScoreData',@params = N' @r_rowsPerRead int, @modeldata varbinary(max)',@r_rowsPerRead = 10000,@modeldata = @model_data;END;GO-- Save predicted labels as tmp tableEXEC GetPredictedResultsAsTable "Decision Tree"-- Merge into original tableUPDATE IrisTblSET IrisTbl.SpeciesPredicted = TempPredicted.SpeciesPredictedFROM IrisTbl INNER JOIN TempPredicted ON IrisTbl.ID = TempPredicted.ID;As you can see below, the predicted values are saved in separated TempPredicted table. Now these results are merged into original table (IrisTbl).
Note : You can also use
rxDataStep()for saving results into SQL table.
As you know, one of pain points with this approach is the difficulty of debugging. You cannot use IDE or utilities for developing R or python code in SQL, then you must debug outside of database and sometimes it might be different from the behaviors in database. Therefore a lot of lines of external script will make you so confused.
With the help of sqlmlutils library, you can easily debug your in-database R script and register your script as an external script.
Note :
sp_execute_external_scriptis now available on Azure SQL Managed Instance.
Pattern 2 : Native Scoring (Built-in T-SQL PREDICT() Function)
If you focus on scoring (predicting) tasks in database, you can also use built-in T-SQL function PREDICT() instead of using sp_execute_external_script. This kind of implementation is called “native scoring” (realtime scoring), because it depends on only SQL Server runtime and does not require external R or python runtime. You can also take a benefit of performance, because it doesn’t use external language runtime. (Especially when it’s called repeatedly.)
One important thing for using native scoring is that you must serialize the trained model with rxSerializeModel() function instead of using as.raw(serialize()). (Since the native scoring will automatically deserialize and distribute the workloads.)
Note : Currently
PREDICT()in SQL Server only supports RevoScale model format, and doesn’t support ONNX model format. (See the below note forPREDICT()model format supports in other SQL database.)
When you need ONNX scoring in SQL Server, you can usesp_execute_external_scriptwith custom external language implementation (in which, ONNX runtime is loaded with extensibility framework) installed. (See here for this implementation.)
See the following highlighted code. (I changed the previous sample code as follows.)
CREATE PROCEDURE GenerateTreeModel ( @model_name VARCHAR(50) OUTPUT, @model_data VARBINARY(MAX) OUTPUT)ASBEGIN EXEC sp_execute_external_script@language = N'R',@script = N'model <- rxDTree( SpeciesLabeled ~ SepalLength + SepalWidth + PetalLength + PetalWidth, data = IrisData, cp = 0.01)modelname <- "Decision Tree"### Change raw format# modeldata <- as.raw(serialize(model, connection=NULL))modeldata <- rxSerializeModel(model, realtimeScoringOnly = TRUE)',@input_data_1 = N'SELECT * FROM IrisTbl WHERE SpeciesPredicted IS NOT NULL',@input_data_1_name = N'IrisData',@params = N' @r_rowsPerRead int, @modelname varchar(50) OUTPUT, @modeldata varbinary(max) OUTPUT',@r_rowsPerRead = 10000,@modelname = @model_name OUTPUT,@modeldata = @model_data OUTPUT;END;
Now the following code is the prediction (scoring) sample with native scoring approach.
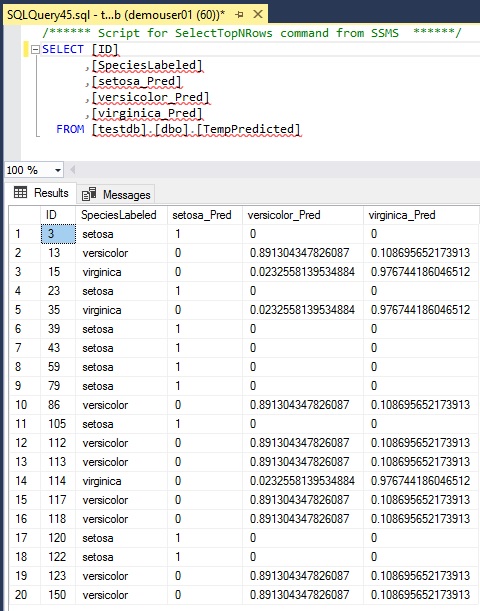
As you can see below, the predicted results are the list of each possibilities with float-type values. (Note that the result is different from the result of rxPredict().)
DROP TABLE IF EXISTS TempPredicted;GOCREATE TABLE TempPredicted ( ID INT, SpeciesLabeled VARCHAR(50), setosa_Pred FLOAT, versicolor_Pred FLOAT, virginica_Pred FLOAT);GODECLARE @model VARBINARY(MAX) = (SELECT ModelData from ModelTbl where ModelName = 'Decision Tree');INSERT INTO TempPredictedSELECT D.ID, D.SpeciesLabeled, P.*FROM PREDICT(MODEL = @model, DATA = IrisTbl AS D) WITH (setosa_Pred FLOAT, versicolor_Pred FLOAT, virginica_Pred FLOAT) AS PWHERE D.SpeciesPredicted IS NULL;GO
Note : You can also use built-in
sp_rxPredictstored procedure (which is the wrapper ofrxPredict()in RevoScaleR) instead. However, the use ofPREDICT()T-SQL function is recommended in SQL Server 2017 or later.
You can also create trained model outside of SQL Server database and bring it within database. For instance, you can train in Spark cluster (Databricks) or Azure Machine Learning, and bring your model for scoring (prediction) in SQL Server.
The following code trains in your working client and save a model in the remote database with Microsoft R Client.
### Read training data from local filecolInfo <- list( list(index = 1, newName="ID", type="integer"), list(index = 2, newName="SepalLength", type="numeric"), list(index = 3, newName="SepalWidth", type="numeric"), list(index = 4, newName="PetalLength", type="numeric"), list(index = 5, newName="PetalWidth", type="numeric"), list(index = 6, newName="SpeciesLabeled", type="factor",levels=c("versicolor", "setosa", "virginica")), list(index = 7, newName="SpeciesPredicted", type="factor",levels=c("versicolor", "setosa", "virginica")))orgData <- RxTextData( fileSystem = "native", file = "C:\\tmp\\iris.csv", colInfo = colInfo, delimiter = ",", firstRowIsColNames = F, stringsAsFactors = T)trainData <- rxDataStep( inData = orgData, rowSelection = !(is.na(SpeciesPredicted)))### Train model !model <- rxDTree( SpeciesLabeled ~ SepalLength + SepalWidth + PetalLength + PetalWidth, data = trainData, cp = 0.01)### Save model to remote databaseserializedmodel <- rxSerializeModel( model, realtimeScoringOnly = TRUE)modelrow <- data.frame(ModelName = 'Decision Tree', ModelData = I(list(serializedmodel)))con <- "Driver=SQL Server;Server=52.170.118.1;Database=testdb;Uid=demouser01;Pwd=P@ssw0rd"modeltbl <- RxSqlServerData( connectionString = con, table = "ModelTbl", rowsPerRead = 10000)rxDataStep( inData = modelrow, outFile = modeltbl, overwrite = TRUE)One important caveat for this approach is that the supported models are limited. Currently the model should be generated by rxLinMod(), rxLogit(), rxBTrees(), rxDtree() or rxDForest() in RevoScaleR (or revoscalepy). Also you cannot bring a model generated by standard R functions (or standard Python functions, such as scikit-learn) for PREDICT() function. (Currently only RevoScale format is supported in native scoring on SQL Server. See above note.)
See the official document “Advanced analytics on SQL Server – Realtime scoring” for details.
Note :
PREDICT()is now available on a variety of SQL-based database family, such as, Azure SQL Database, Azure SQL Managed Instance, Azure SQL Edge, and Azure Synapse Analytics.
However, the supported model format differs between each SQL-based database engines. See here for the currently supported format in each database.
Pattern 3 : Attach SQL Server Compute Context From Remote (RxInSqlServer)
The last approach is a bit different from previous two approaches.
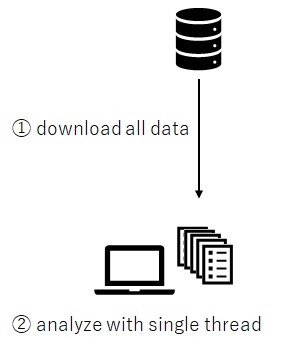
Now let’s consider the following code for remote access with ODBC module.
library(RODBC)library(tree)con <- "Driver=SQL Server;Server=52.170.118.1;Database=testdb;Uid=demouser01;Pwd=P@ssw0rd"ch <- odbcDriverConnect(connection = con)### Traintrainds <- sqlQuery(ch, "SELECT * FROM IrisTbl WHERE SpeciesPredicted IS NOT NULL")model <- tree( SpeciesLabeled ~ SepalLength + SepalWidth + PetalLength + PetalWidth, data = trainds)### Predicttestds <- sqlQuery(ch, "SELECT * FROM IrisTbl WHERE SpeciesPredicted IS NULL")labels <- predict(model, testds, type='class')
Note : Be sure to open tcp port 1433 in firewall settings.
As you know, all data is transferred into the client and processed by the single thread in your client. If the size of data is so large, it will cause latency for data IO.
In such a case, you can also take benefits of in-database workloads by using SQL Server compute context.
Now let’s see the next example.
### Set compute contextcon <- "Driver=SQL Server;Server=52.170.118.1;Database=testdb;Uid=demouser01;Pwd=P@ssw0rd"sqlCompute <- RxInSqlServer( connectionString = con, wait = T, numTasks = 5, consoleOutput = F)rxSetComputeContext(sqlCompute)#rxSetComputeContext("local") # if debugging in local !### Traintrainds <- RxSqlServerData( connectionString = con, databaseName = "testdb", sqlQuery = "SELECT * FROM IrisTbl WHERE SpeciesPredicted IS NOT NULL", rowsPerRead = 10000, stringsAsFactors = T)### Retrieve and show remote data# test <- rxGetInfo(# data = trainds,# numRows = 150)# test$datamodel <- rxDTree( SpeciesLabeled ~ SepalLength + SepalWidth + PetalLength + PetalWidth, data = trainds, cp = 0.01, minSplit = sqrt(130), # data size is 130 maxNumBins = min(1001, max(101, sqrt(130)))) # data size is 130### View result tree with browser# library(RevoTreeView)# plot(createTreeView(model))### Predicttestds <- RxSqlServerData( connectionString = con, databaseName = "testdb", sqlQuery = "SELECT * FROM IrisTbl WHERE SpeciesPredicted IS NULL", rowsPerRead = 10000, stringsAsFactors = T)sqlServerOutDS <- RxSqlServerData( connectionString = con, table = "PredTbl", rowsPerRead = 10000)rxPredict( model, data = testds, type = "class", outData = sqlServerOutDS, writeModelVars = TRUE, overwrite = TRUE, predVarNames = "SpeciesPredicted")The logic flow of this code is similar to the previous one (ODBC sample). But this code works so different from the previous one.
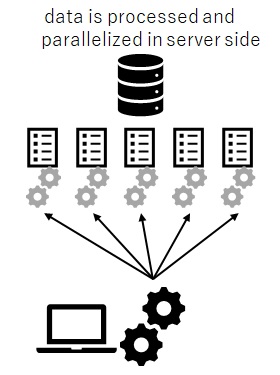
Same as RevoScaleR functions in Machine Learning Server (see “Benefits of Microsoft R and Machine Learning Server“), the data is not transferred across network. The machine learning tasks will be run and parallelized on the server side (close to data) by using RxInSqlServer context. (See the following illustrated picture.)
That is, you can take advantages of reducing overhead of network transfer, streaming IO of data, and multiple processes by using RxInSqlServer context. (In order to take advantages for streaming and multiple processes, you must use SQL Server Enterprise, as I mentioned above.)
Note that I’m setting concurrent process (numTasks attribute) as 5 in above example, but the database will determine the actual concurrent number depending on the server resources. (In this case, our sample data is so small, and less processes might be used.)
This approach makes debugging so easy. However, one pain point is that you must care about several tricky things, because data is not in local machine.
For example, you cannot use standard R functions (rpart(), tree(), so on and so forth) for data manipulation with RxInSqlServer context. (You should use RevoScaleR or revoscalepy functions instead.)
You cannot also have predicted-results as local objects, such as files or memory data. Therefore you should save the results into SQL table by setting outData attribute as above.
You must also explicitly specify minSplit and maxNumBins attributes in rxDTree() as above, because the number of data is unknown.
Note : SQL Server compute context running on Linux is supported in ML Server 9.3 release.
When you want to run some standard R blocks (without RevoScaleR functions), you must write as follows using rxDataStep() . This code reads data in chunk and applies R functions to each chunk in turn.
processfunc <- function(df) { ... # write standard R block running in server side return(...)}OutputDataSet <- rxDataStep( inData = IrisData, outFile = tempResult, transformFunc = processfunc, overwrite = T)Note : Remote compute context is now available only for on-premise SQL Server. (You cannot use for other SQL database.)
As you saw in this post, in-database workloads is so scalable and you can handle tens of millions of data for machine learning.
In the next post, I’ll show you how to run deep learning (AI) workloads on powerful SQL Server ML Services.
Categories: Uncategorized

Hi Tsuyoshi,
When installing SQL 2017 instance without the ”Advanced Analytics Extensions”, I’m not able to use sp_execute_external_script from SSMS. That is expected.
However, when I’m trying to use RevoScaleR from my RStudio locally, I can have the remote compute context established as below. My question is without the “Advanced Analytics Extensions” installed remotely, is RevoScaleR still using the remote SQL Server as the compute context? I thought it won’t have the R runtime required. Is there a simple example I can test which context is used?
> rxGetComputeContext()
RevoScaleR Microsoft SQL Server In-Database Object
————————————————–
connectionString : “Driver=SQL Server;Server=XXXXX;Database=test_Dream;Trusted_Connection=True;”
numTasks : 1
executeAsUser : “”
executionTimeoutSeconds : 0
shareDir : “C:\\AllShare\\XXX”
wait : TRUE
consoleOutput : FALSE
autoCleanup : TRUE
nodes : NULL
packagesToLoad : NULL
description : “sqlserver”
version : “1.0-2”
LikeLike